We are happy to share that 3 pre-prints reporting exciting new results by the FOR2581 members are now available:
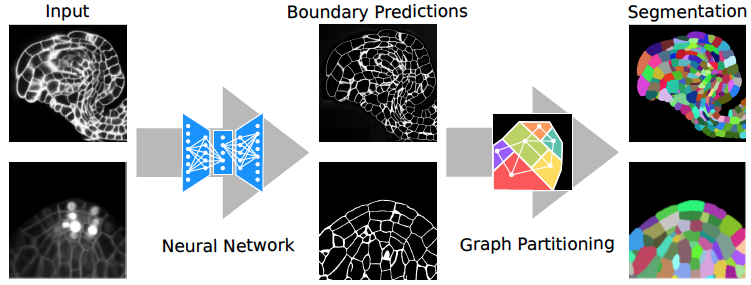
Our joint effort to develop a method to segment plant cells in tissue are paying off. The combined use of neural networks and graph partitioning algorithms deliver accurate, efficient and versatile results!
> Wolny, A., Cerrone, L., Vijayan, A., Tofanelli, R., Barro, A.V., Louveaux, M., Wenzl, C., Steigleder, S., Pape, C., Bailoni, A., et al. (2020). Accurate And Versatile 3D Segmentation Of Plant Tissues At Cellular Resolution. BioRxiv
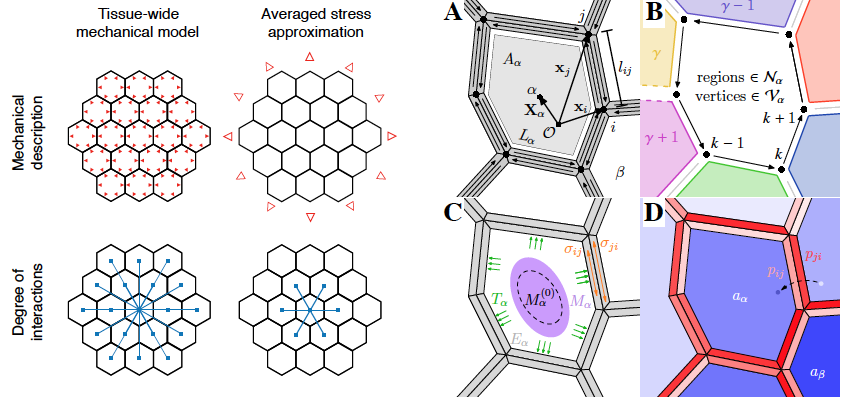
A therotical work from the Alim (and Maizel) labs that compares a model accounting for a tissue-wide mechanical integration to a model where mechanical stresses are averaged out across the tissue. They show that only tissue-wide mechanical coupling leads to focused auxin spots.
>Ramos, J. R. D., Maizel, A., Alim, K. (2019) Tissue-wide integration of mechanical cues promotes efficiency of auxin patterning. BioRxiv
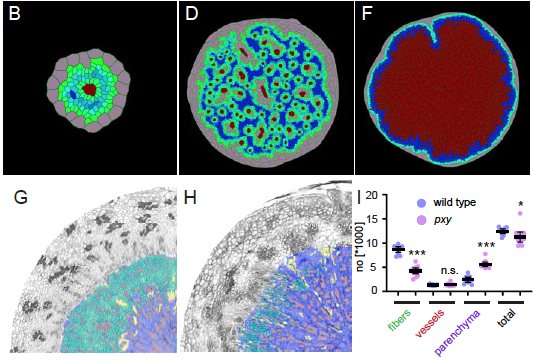
The Greb lab, in collaboration with the Merks (Leiden NL) used modelling to shows that a limited number of factors is sufficient to create a stable bidirectional tissue production in the radially expanding hypocotyl.
Lebovka, I., Mele, B.H., Zakieva, A., Gursanscky, N., Merks, R.M.H., and Greb, T. (2020). Computational modelling of cambium activity provides a regulatory framework for simulating radial plant growth. BioRxiv
For a complete list, see the Publications section.